Programme
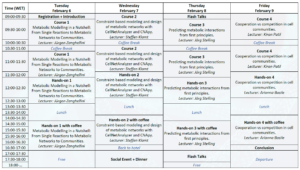
Course 1 and hands-on 1 on Feb 6 will be given by Prof. Jürgen Zanghellini
Course 2 and hands-on 2 on Feb 7 will be given by Prof. Steffen Klamt
Course 3 and hands-on 3 on Feb 8 will be given by Prof. Jörg Stelling and Constance Le Gac
Course 4 and hands-on 4 on Feb 9 will be given by Prof. Kiran Patil. Hands-on 4 will also be given by Arianna Basile
Lecturers





Prof. Jürgen Zanghellini, University of Vienna, Austria
Title: Metabolic Modelling in a Nutshell: From Single Reactions to Metabolic Networks to Communities.
Abstract: This primer provides an introduction to metabolic modeling, guiding participants through key concepts. It starts with an exploration of collision theory, offering a physical understanding of biochemical reactions. The discussion extends to deriving standard enzyme kinetics and applying them to model dynamic features of metabolism. Participants will also delve into network reconstruction and general growth models to enhance their insights. During the practical exercises, Copasi will be used for dynamic analysis, while COBRApy (https://doi.org/10.1186/1752-0509-7-74) and PyCoMo will be employed for manipulating genome-scale metabolic models and exploring community formation and characterization in steady state. To run the latter analysis I will use Jupyter notebooks.
Biosketch: Jürgen Zanghellini is a Professor of Chemical Bioinformatics Network Analysis at the Faculty of Chemistry, University of Vienna, Austria. He completed his studies in Electrical Engineering (2001) and earned a Ph.D. in Theoretical Physics (2004), both at the Technical University of Vienna. His research focuses on mathematically modeling the dynamics, regulation, and control of metabolic networks. Specifically, he explores the structural, i.e., topological properties of complex metabolic networks and investigates how these structures contribute to metabolic functions. Dr. Zanghellini and his team develop computational tools to predict optimized microbes for various biotechnological applications. Publications here (Link 1, Link 2).
Prof. Steffen Klamt, Max Planck Institute for Dynamics of Complex Technical Systems, Germany
Title: Constraint-based modeling and design of metabolic networks with CellNetAnalyzer and CNApy.
Abstract: This 1-day course introduces principles of constraint-based modeling and computational design of metabolic networks coupled with a live demonstration and hands-on exercises with the MATLAB package CellNetAnalyzer (CNA) and its Python-variant CNApy. The lecture will overview the key methods of constraint-based modeling and metabolic network design:
– Fundamentals of constraint-based modeling of metabolic networks
– Metabolic flux (balance) analysis
– Flux Variability analysis
– Phase planes and Yield spaces
– Metabolic pathway analysis
– Minimal cut sets and computational design of metabolic networks.
In the Hands-on Session the participants will have a guided tour through key features of CellNetAnalyzer (CNA) and its Python-variant CNApy (live demonstration) and will practice the use of CNA or/and CNApy by exercises with small example networks as well as with realistic metabolic models.
Requirements for hands-on exercises:
Please install/download CNA or/and CNApy prior to the course and test whether it runs on your system (you should have installed at least one of the two toolboxes). You can download the latest version of CNA from the following website:
https://www2.mpi-magdeburg.mpg.de/projects/cna/cnadownload.html
For using CNA you need MATLAB. If you do not have access to MATLAB you may download a 30 day trial version here:
https://www.mathworks.com/campaigns/products/trials.html
For installation of CNApy please follow the instructions on:
https://github.com/cnapy-org/CNApy
Please download also the CNApy example projects from:
https://github.com/cnapy-org/CNApy-projects
Finally, for the hands-on exercises please aslo download the following two files:
https://www2.mpi-magdeburg.mpg.de/projects/cna/exercise.pcx
https://www2.mpi-magdeburg.mpg.de/projects/cna/exercise.svg
The slides of the course can be downloaded here.
Biosketch: Dr. Steffen Klamt heads the group “Analysis and Redesign of Biological Networks” at the Max Planck Institute for Dynamics of Complex Technical Systems in Magdeburg, Germany. He received his Diploma in Systems Science from the Osnabrück University in 1998 and his Ph.D. from the Stuttgart University in 2005. His group develops methods for systems and computational biology and combines them with experimental investigations. One central research focus is modeling and computational design of metabolic networks with applications in metabolic and biosystems engineering. His group developed CellNetAnalyzer and CNApy, two comprehensive packages for the in silico analysis and rational design of biochemical networks. Experimental work focuses on Escherichia coli as model organism and tight integration of wet-lab and dry-lab investigations is used to verify model-based predictions and to run Design-Build-Test-Learn cycles of metabolic engineering. Steffen Klamt has co-authored over 100 peer-reviewed publications and is editorial board member of BMC Bioinformatics and Biotechnology Journal.
German Network for Bioinformatics Infrastructure.
Prof. Jörg Stelling, ETH Zürich, Switzerland
Title: Predicting metabolic interactions from first principles.
Abstract: In modeling interactions between cells or in communities, a challenge is to predict potential or actual metabolic exchanges. Traditional constraint-based modeling methods such as flux balance analysis require additional assumptions (such as an objective for the community as a whole) or knowledge on kinetics that is often not available (such as in dynamic flux balance analysis). The lecture will give an overview of more recent methods that aim to address this limitation. They integrate first principles (e.g. thermodynamics or enzyme costs) into constraint-based models, or rely on metabolic pathway analysis and game theory approaches.
Requirements for hands-on exercises:
In the hands-on sessions, participants will explore corresponding tools with practical examples, starting from the method of probabilistic thermodynamic analysis (PTA, https://academic.oup.com/bioinformatics/article/37/18/2938/6182675) and the estimation of enzyme kinetic parameters (ENKIE, https://www.biorxiv.org/content/10.1101/2023.03.08.531697v1).
Requirements for hands-on-sessions: Please prepare your computer such that the requirements for installing PTA are fulfilled (section 1.1, including the optional components at https://probabilistic-thermodynamic-analysis.readthedocs.io/en/latest/getting_started.html), but do NOT yet install PTA. In particular, install the Gurobi solver using the free (for academic use) personal or WLS license.
In addition, install an application for running Jupyter notebooks, such as JupyterLab (https://jupyter.org/install) or corresponding functionalities for Visual Studio Code (https://code.visualstudio.com/; https://marketplace.visualstudio.com/items?itemName=ms-toolsai.jupyter). Finally, you need to have a working installation of R (see e.g. here, if you do not have one yet: https://rstudio-education.github.io/hopr/starting.html).
Biosketch: Jörg Stelling is Professor of Computational Systems Biology in the Department of Biosystems Science and Engineering (D-BSSE) at ETH Zurich. After receiving his Ph.D. in Control and Dynamic Systems from Stuttgart University in 2004, he joined the Computer Science Department at ETH Zurich, and in 2008 the newly founded D-BSSE. Jörg Stelling works at the interface of systems and synthetic biology, using – and further developing – methods from systems theory and computational science. Main aims of current research are to understand how inter-individual variability affects the functioning of complex networks, to expand constraint-based modeling methods for the analysis of microbial communities, and to provide concepts and methods for the rational design of biological systems despite uncertain and non-robust components and interactions.
Prof. Kiran Raosaheb Patil, MRC Toxicology Unit and Department of Biochemistry, University of Cambridge, UK
Title: Cooperation vs competition in cell communities.
Abstract: Multi-cellular biological systems – from microbiomes to tumours to human body – need to function under resource limitation. Yet, dependencies between cells have evolved leading to fascinating but intricate networks. I will discuss how metabolic modelling can be used to unravel competitive and cooperative forces operational in cell communities and applications thereof.
Biosketch: Kiran studied Chemical Engineering at the Indian Institute of Technology (Mumbai, India). Following his PhD in Systems Biology at the Technical University of Denmark (DTU), Kiran was appointed as Assistant Professor at DTU where he worked on transcriptional regulation and metabolic engineering. In 2010, Kiran was appointed Group Leader at the European Molecular Biology Laboratory (Heidelberg, Germany). He joined MRC Toxicology Unit (University of Cambridge) as Director of Research in 2019 and Department of Biochemistry in 2020. His lab develops tools and model systems to decipher and modulate metabolic interactions.
Dr. Arianna Basile, MRC Toxicology Unit and Department of Biochemistry, University of Cambridge, UK
Hands-on Course 4
Biosketch: Arianna Basile has a background in Bioinformatics and obtained her PhD in Bioscience (genetics, genomics and bioinformatics) at University of Padova (Italy) in 2022 with Professor Giorgio Valle. During her PhD, Arianna developed tools and applications in the context of Flux Balance Analysis applied to the study of microbial anaerobic metabolism (i.e. anaerobic digestion and human gut flora). In 2020, Arianna was awarded an European short-term fellowship from EMBO, and she flied in Ireland where she had the honour to work with Professor Ines Thiele and Professor Almut Heinken. In 2022, Arianna joined Kiran Patil’s lab at the MRC Toxicology Unit.
Requirements for hands-on exercises:
https://github.com/arianccbasile/FBA_OLISSIPO_Winter_School/tree/main. The requirements are specified in the repository and are the following: cobra, matplotlib, numpy, micom, pandas, os, jupyter notebook.
Registration
Registration is free but mandatory for organizational purposes. Seating is limited and on a first-come, first-served basis. Please register here (https://forms.gle/HBVK9vpLoEEjxJ7V8) until January 12, 2024.
Lunches from Tuesday to Friday are included, accommodation and flights are not.
The social event and dinner on Wednesday have limited seats (requires confirmation from the organisation).
Participants
The school is open to all members of the OLISSIPO project but also, importantly, to the community in general. National and international PhD students and researchers studying or interested in studying Mathematical modelling of inter-cell and communities’ interactions with a focus on metabolism are therefore welcome.
Flash Talks
We have slots for flash talks (3-10 min) by ESRs to present the work they have been developing in their research.
Organisers
Susana Vinga (INESC-ID), Marie-France Sagot (Inria), Niko Beerenwinkel (ETH Zürich), Wolfgang Huber (EMBL), and Sara Tanqueiro (INESC-ID)
Funding
The H2020 Twinning Project Olissipo has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 951970.


