Keynote Speakers

Ana Conesa, Institute for Integrative Systems Biology, Spanish National Research Council
Title Talks: Long Reads sequencing to unravel the functional impact of alternative splicing.
Multiomics tools to model dynamics cellular systems in health and disease.
Biosketch: Ana Conesa is a Research Professor at the Institute for Integrative Systems Biology (CSIC) in Valencia (Spain) and Courtesy Professor at the University of Florida. She graduated as Agricultural Engineer at the Polytechnical University of Valencia in 1993 and did her PhD in at the University of Leiden in the Netherlands. After a short appointment as bioinformatics project leader at TNO Quality of Life (The Netherlands) she obtained a Ramon y Cajal award and joined the Valencia Agricultural Research Institute in 2003. She held positions as group leader at the Principe Felipe Research Center (2007-2018), and as full professor at the University of Florida (2014-2020). In 2022 she became member of the Spanish Royal Academy of Engineer and of the Board of Directors of the International Society for Computational Biology.
Ana Conesa’s lab is interested in understanding functional aspects of gene expression at the genome-wide level and across different organisms. Her group has developed statistical methods and software tools that analyze the dynamics aspects transcriptomes, integrate these with other types of molecular data and annotate them functionally, with a special focus on Next Generation Sequencing (NGS) data. A strong drive in her research is helping the genomics community to bridge the gap between data and knowledge by creating bioinformatics tools that everybody can use. Some of our popular software tools are Blast2GO, PaintOmics, maSigPro, NOISeq, Qualimap, SQANTI, tappAS, etc. She has led large international projects such as STATegra and DEANN where European and American scientists developed new tools for the analysis of sequencing data and is co-organizer of LRGASP, a community-wide contest to assess the utilization of long reads sequencing for transcriptome research. She is organizer of major international conferences in Computational Biology, has delivered over 50 keynote lectures and regularly acts as scientific advisor for funding agencies, research institutions and companies world-wide. She is also co-founder of Biobam Bioinformatics, a start-up that provides bioinformatics tools for biologists. She has published 155 research papers that have received more than 30.000 citations and has an h-index of 55.

Laura Elo, University of Turku
Title Talk: Computational tools to study transcriptional landscapes with single-cell resolution.
Abstract: We provided an overview of computational tools used to analyse single-cell transcriptomics data. Single-cell transcriptomics is a rapidly growing field that allows researchers to study gene expression at the level of individual cells, providing insight into cellular heterogeneity and function. The course covered the basics of single-cell transcriptomics data processing and introduced participants to various computational tools and methods for analysing the data, including clustering, differential gene expression analysis, and trajectory inference. We also briefly discussed recent advances in single-cell multi-omics techniques to complement the transcriptomics analysis. Throughout the course, we used real-world examples to illustrate the concepts and methods covered in the lectures. With a combination of lectures and hands-on analysis, the participants learnt how to apply the computational tools using the R programming language to study transcriptional landscapes with single-cell resolution and gain a better understanding of the underlying biological processes.
Biosketch: Laura Elo, PhD, is Professor of Computational Medicine and Head of Medical Bioinformatics Centre at University of Turku, Finland. She has PhD in applied mathematics and long experience in molecular systems immunology and application of machine learning in medical research in several research projects (e.g. ERC). Her research group (https://elolab.utu.fi) develops statistical and machine learning methods to interpret large-scale molecular and clinical data, including single-cell data and their integration. She has published >150 research articles and >20 software packages.

Sini Junttila, University of Turku
Title Talk: Analysing single-cell data using R.
Participants should install R and certain packages on your laptops.
The requirements are:
- R >= 4.2.0 (https://www.r-project.org)
- CRAN packages: devtools, Seurat, dplyr, patchwork
- Bioconductor packages (https://www.bioconductor.org): ROTS, edgeR, scuttle, SingleR, celldex
- Github package: Totem, https://github.com/elolab/Totem
The installation of RStudio (https://posit.co) is highly recommended, but not compulsory.
Biosketch: Sini Junttila, PhD, is a post-doctoral researcher at the Medical Bioinformatics Centre at University of Turku, Finland. Working several years at core facilities in Finland and Austria she has gained extensive experience in analysing a wide variety of high-throughput omics data. Her main research areas are epigenomics and single-cell RNA-sequencing. Recently she has been developing and benchmarking tools for single-cell RNA-seq analysis.

Mathurin Massias, Inria Lyon
Title Talks: Theoretical and practical aspects of dimensionality reduction.
Abstract: Dimensionality reduction (DR) is of central importance when dealing with high-dimensional datasets and, with the current explosion of the data size, DR is at the heart of many problems in data science. In bioinformatics, for example, genetic data often involve very large vectors, making it difficult to learn, interpret or explore such data. In this context, DR enables visualization that can be of great practical interest for understanding and interpreting the structure of large datasets. DR also mitigates the curse of dimensionality, allowing for greater statistical flexibility and less computational burden. In this course, we introduced most popular DR methods both from a practical and a theoretical point of view. More precisely this course covered linear DR methods such as principal component analysis, random projections, spectral embedding and non-linear methods such as SNE, t-SNE and deep autoencoders. The class was complemented by two practical labs in Python. They leveraged the `scikit-learn` and `pytorch` Python packages to highlight the various benefits and possible behaviors of the presented methods. Experiments and visualisation on real genomics data demonstrated the interest of using DR techniques.
Prerequisite: Basics of Python programming: https://swcarpentry.github.io/python-novice-inflammation; Introduction to linear algebra for Machine Learning: https://pabloinsente.github.io/intro-linear-algebra (and references therein).
Other useful references:
[1] Laurens Van Der Maaten, Eric Postma and Jaap Van den Herik, ***Dimensionality Reduction: A Comparative Review***, Journal of Machine Learning Reseach, 2009.
[2] Akshay Agrawal, Alnur Ali and Stephen Boyd, ***Minimum Distortion Embedding***, Foundations and Trends® in Machine Learning, 2021.
Participants should have a working installation of python with jupyter notebooks. We recommend installing python through Anaconda for simplicity. We will rely on classical packages from the python ecosystem: numpy, scipy, matplotlib, pandas, scikit-learn, pytorch.
Biosketch: Mathurin Massias is a permanent researcher (chargé de recherche) at INRIA Lyon, working on large scale optimization for Machine Learning. He develops new optimizations methods that allow treating the datasets of evergrowing size that appear in many applied sciences. He obtained his PhD from Inria Saclay in 2019, on fast methods for high dimensional sparse regression with applications to neural source localization with magneto-electroencephalography. He currently works on new efficient algorithms for sparse learning in high dimension, and non convex problems arising in deep learning.

Titouan Vayer, Inria Lyon
Title Talks: Theoretical and practical aspects of dimensionality reduction.
Biosketch: Titouan Vayer is a permanent researcher (chargé de recherche) at INRIA Lyon and works on compressive learning problems, i.e. on how to reduce the dimension of the data while guaranteeing good learning performance. He worked during his thesis, which he obtained in 2020 in IRISA, Vannes, on optimal transport methods for machine learning, in particular for graphs and heterogeneous data. Titouan Vayer is particularly interested in the theory of learning in complex settings where the data are large, structured and do not admit the same representation.

Stefan Canzar, University of Munich
Title Talk: Engineered algorithms for large-scale single-cell RNA sequencing and multimodal data analysis.
Abstract: Experimental methods for sequencing DNA or RNA of single cells have transformed biological and medical research. The throughput of this technology has dramatically increased over the last few years, such that today the expression of genes in millions of cells can be measured in a single experiment. The computational interpretation of the produced data, however, often exceed the capacity of existing algorithms. In practice, therefore, scRNA-seq analysis methods are often run on a random sample of cells or take into account only a subset of the constraints that define the computational task. Simply ignoring large parts of the experimental measurements, however, can lead to inaccurate solutions and wrong biological conclusions, rendering experimental advances meaningless. In the first part of this course, we described an efficient algorithm that uses techniques for the design of approximation algorithms to compute a small subset of cells that represent the transcriptional space of the original data as accurately as possible. The resulting sketch of single cells facilitates visualization and sharing of large datasets and accelerates downstream analyses such as clustering while retaining high accuracy. In addition, we explained how marker genes that discriminate cell types can be selected by imposing distance constraints on all pairs of cells from different types, using large-scale optimization techniques to avoid explicitly generating this massive number of constraints. Emerging single-cell technologies allow to measure the abundance of different types of molecules in individual cells, including the transcriptome, genome, epigenome, and the proteome. The second part of this course covered computational methods that combine measurements of multiple types of molecules such as the expression of mRNAs and surface proteins to refine cell types. Furthermore, we describe an extension of t-SNE and UMAP, the most popular methods for the visualization of high-dimensional biomedical data, to the joint visualization of such modalities.
Biosketch: Stefan Canzar received his Diploma in Computer Science from the Technical University of Munich, Germany, in 2004, and his bi-national doctoral degree in Computer Science from Saarland University, Germany, and Henri Poincaré University, Nancy, France, in 2008. From 2009 to 2011 and from 2012 to 2014, he was a post-doctoral researcher at the Centrum Wiskunde & Informatica, Amsterdam, The Netherlands, and the Johns Hopkins Institute of Genetic Medicine, Baltimore, USA, respectively. From 2014-2016, he was a Research Assistant Professor with the Toyota Technological Insitute at Chicago. Since 2016 he is a Research Group Leader at the Gene Center of the University of Munich (LMU) and will join The Pennsylvania State University as Associate Professor in January 2023. His research interests include the development of algorithmic solutions to problems arising in the analysis of high-throughput sequencing data. Stefan Canzar and his group have developed efficient algorithms that deal with the complexity and scale of single-cell multi-omics data. The Canzar lab has translated theoretical insights into practical, open-source software tools that it uses in collaborations to address a wide range of biological and medical questions.
Program
This school had two main types of presentations: one that had a class format exclusively, and the other that had a class plus hands-on session format. The class was divided into two slots of 1h and 1h30. The same happened for the hands-on session.
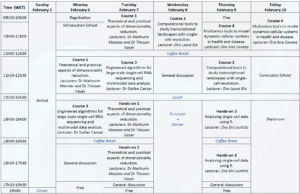
Registration
Registrations are closed!
The registration fee was 350€ (cocktail + dinner on day 5, all lunches + coffee breaks and the excursion + dinner on day 8 are included, accommodation and flights are not).
Participants
The school was open to all members of the OLISSIPO project but also, importantly, to the community in general.
Hotel suggestions
The Olissippo Marquês de Sá hotel, the venue where the school was held, made a special price for the participants of this OLISSIPO event between February 5-10, 2023. After registration, you will receive an e-mail with the promotional code to benefit from this advantage by booking your accommodation at this link: https://www.olissippohotels.com/pt/Hoteis/Marques-de-Sa/O-Hotel.aspx:
There are also many hotels and holiday rental options in Lisbon, check the usual websites (Booking, trivago, tripadvisor, airbnb, …).
Photos
The H2020 Twinning Project Olissipo has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement No 951970.











