OLISSIPO School | EMBL Training

The EMBL team, one of the partner institutions of the OLISSIPO project, visited INESC-ID to give a series of workshops in the area of Computational Biology.
Location: INESC-ID (Lisboa, Portugal), Room 9
Agenda:
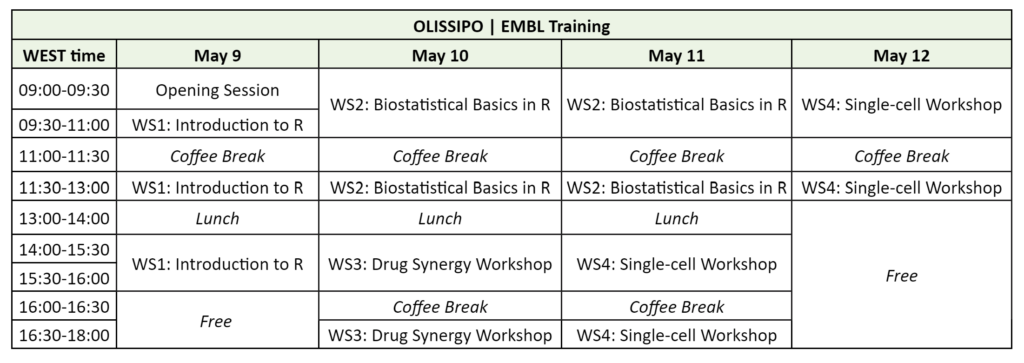
Workshop 1 (WS1) (May 9, 09:00-16:00)
Title: Introduction to R
Lecturer: Mike Smith
This one day course provided an introduction to data handling with the R language and covered some useful tools for data wrangling, exploration and plotting using the tidyverse. The course material was suitable for beginners, although basic familiarity with R was advantageous.
Prerequisites: A working installation of R and RStudio. Installation instructions for both can be found under “Setup Instruction” on https://grp-bio-it-workshops.embl-community.io/introduction-to-R/index.html
Workshop 2 (WS2) (May 10-11, 09:00-13:00)
To attend this course it was necessary to attend Workshop 1
Title: Biostatistical Basics in R
Lecturer: Sarah Kaspar
This course equipped participants with basic statistical concepts that helped them to better understand their biological data, and which are also the foundation for many advanced biostatistical topics.
We covered:
- statistical distributions
- hypothesis testing
- multiple testing
- contingency tables
Prerequisites: Participants needed to have R and RStudio installed, and basic knowledge of R and data visualisation using the tidyverse. Dor those who didn’t have this pre-knowledge they needed to complete one of the following:
- Tuesday’s “Introduction to R” course
- The carpentries Introduction to R (working on your local RStudio)
- The EMBL-EBI tutorial on data handling and visualisation in R (working in the browser)
Workshop 3 (WS3) (May 10, 14:00-18:00)
Title: Drug Synergy
Lecturer: Petr Smirnov
This workshop introduced the fundamental concepts behind analysing drug combination experiments on immortalised cancer cell lines; discussed common models for assessing drug synergy, and introduced R/Bioconductor tools implementing these methods. A hands on analysing public drug combination data was included.
The learning goals behind this workshop were:
- Describe pharmacogenomic mono and combination datasets and usefulness in cancer research
- Understand how experimental designs and research questions map onto data structures
- Learn how to extract information from these datasets
- Learn how to visualise experimental results from these datasets
- Learn how to model dose-response for both monotherapy and combination small compound datasets
- Learn measures to quantify response and synergy in cell line sensitivity screens
Prerequisites: Participants needed working knowledge of R and a basic familiarity with Bioconductor. Experience with the data.table R package was helpful, but not required.
Workshop 4 (WS4) (May 11, 14:00-18:00; May 12, 09:00-13:00)
Title: Single-Cell
Lecturers: Donnacha Fitzgerald, Hosna Baniadam, Harald Vöhringer, Petr Smirnov and Tümay Capraz
The aim of this workshop was to introduce single-cell (including multi-omic and spatial) methods, what can be achieved with them and principles for their analysis. It included the following topics:
- Overview of single-cell methods
- Single-cell transcriptomics analysis
- Single-cell data integration (horizontal and vertical)
- Spatial analysis + probabilistic factor models for the analysis of single cell data
Each topic included a 30 minutes talk followed by a 45 minutes practical component.
We also did a consultation hour (60 minutes) at the end of the program for people to ask questions related to their own projects or to bring data for help with their analysis.
Registration was free (30 seats for each workshop).
We would like to thank Donnacha, Harald, Hosna, Mike, Petr, Sarah and Tümay, our wonderful lecturers from the OLISSIPO partner EMBL for accepting this invitation to help us train our Early-Stage Researchers.




